FAQs¶
If you have questions, requests, or bugs to report, please use the Metaboverse issues forum.
Reporting problems during model building¶
If you encounter an error during the build of your model, you should see a notification alerting you that a error log has been output. This file will be output to your selected
output folder. Error logs are output to ensure all relevant details are visible to the user and developers. You may read through the log to see if there is a solution you can find, or you can submit an issue to us at the Metaboverse issues forum and attach the log file there for us to review.Model building not showing any progress¶
Most likely, if Metaboverse does not show any progress on the
Build page, this is due to a permissions issue. This can often be remedied by running Metaboverse as an administrator. Do so by right-clicking on the Metaboverse app icon and select Run as administrator. If a error log is output, please submit it to the Metaboverse issues forum.Model building freezes¶
A frozen progress bar during model building is often due to internet connection issues. Please check you internet connection and connection speed in case this is the issue. If your internet connection seems fine, please submit an issue to the Metaboverse issues forum.
Not accepting a particular input¶
Try substituting spaces with underscores in input fields. Please also attach any errors or logs you receive when this happens and report on the Metaboverse issues forum.
Linux app not launching¶
If you click on the Metaboverse app for Linux and you see the following error:
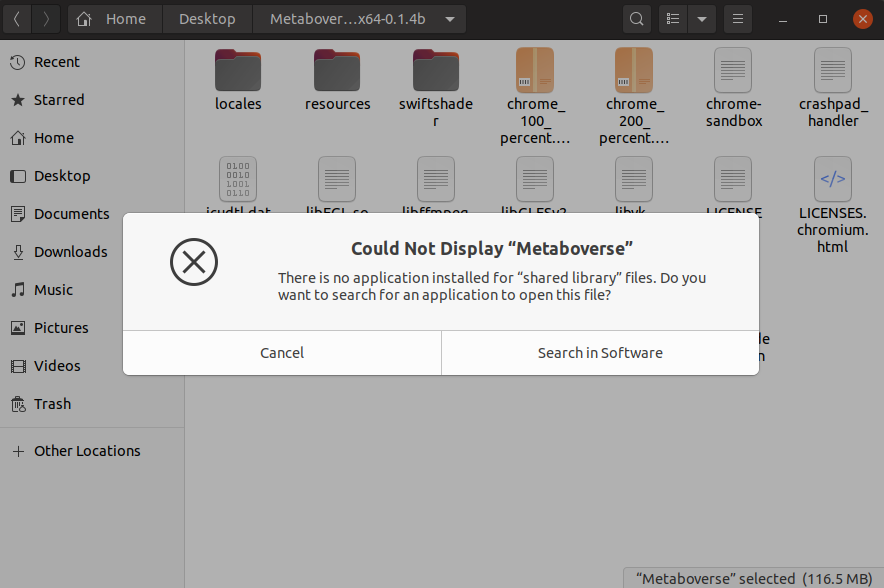
you should perform the following steps in the Terminal.
$ cd /path/to/unzipped/metaboverse/app/folder
$ chmod +x ./Metaboverse
$ chmod +x ./resources/app/python/metaboverse-cli-linux
and then launch the app by executing the following:
$ ./Metaboverse
Error: AttributeError: 'float' object has no attribute 'lstrip'¶
If the error,
AttributeError: 'float' object has no attribute 'lstrip', appears this is likely caused by missing values in one of the provided data tables. While current procedures should now handle this issue, if this error persists, you might try deleting any trailing whitespace from the gene/protein/metabolite names in your data tables.